After a long time and so many commits (more than 350!), it is now time to publish a new release.
Changelog is quite long but includes:
- Build for MacOS X Big Sur and AppImage format for Linux build
- Allow import of metadata from an Excel or ODF spreadsheet
- Allow to remove group mappings columns and update mappings when loading mapping file instead replacing everything
- Delay import dependencies modules for workers (faster loading of main interface)
- Scores matrix is loaded only when needed instead at project opening (faster project opening)
- Allow to copy molecular structure to copy clipboard as images from Database dialog
- Export of Database Results in CSV format
- Allow to export a subset of metadata
- Add a warning if user tries to import an empty data file
- Allow to copy molecular structure to copy clipboard as images from Database dialog
- Export of Database Results in CSV format
- Mechanism to check and notify user for new versions on start-up
- Improved SVG export and add EMF export for network views
Many User Interface improvements have been made, including:
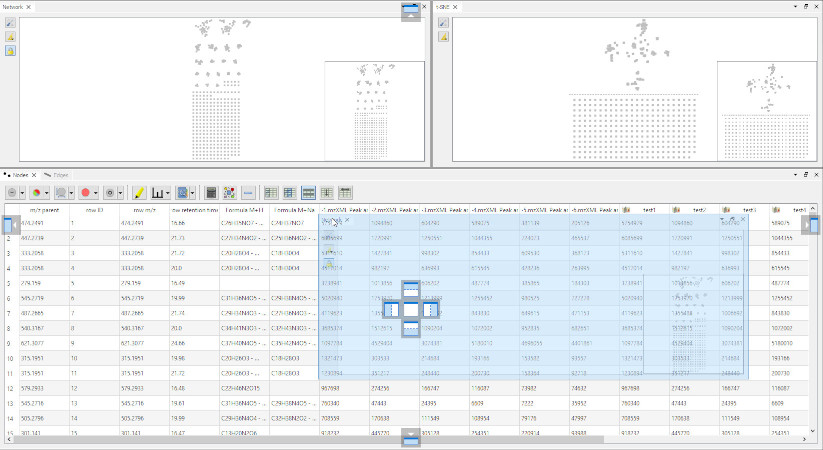
- Use of Qt-Advanced-Docking-System to manage docks and allow to create network views independently. It is possible to create more than one view of each type (multiple t-SNE for example):
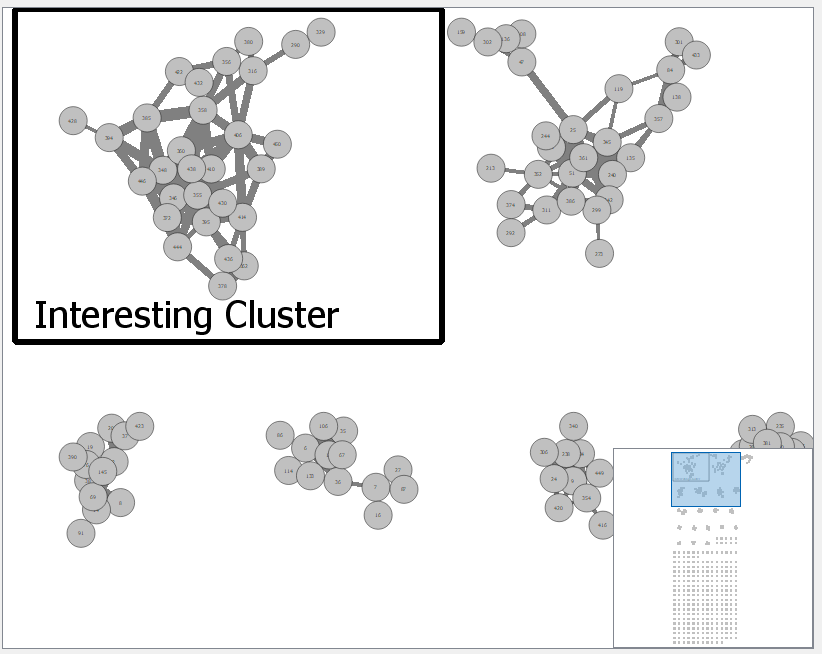
- Network annotations:
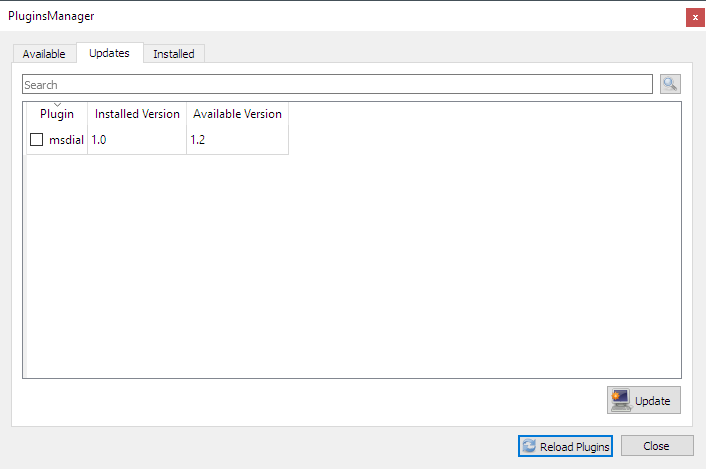
- A plugin mechanism for database downloading has been developed. Plugins can be updated independently directly from MetGem interface:
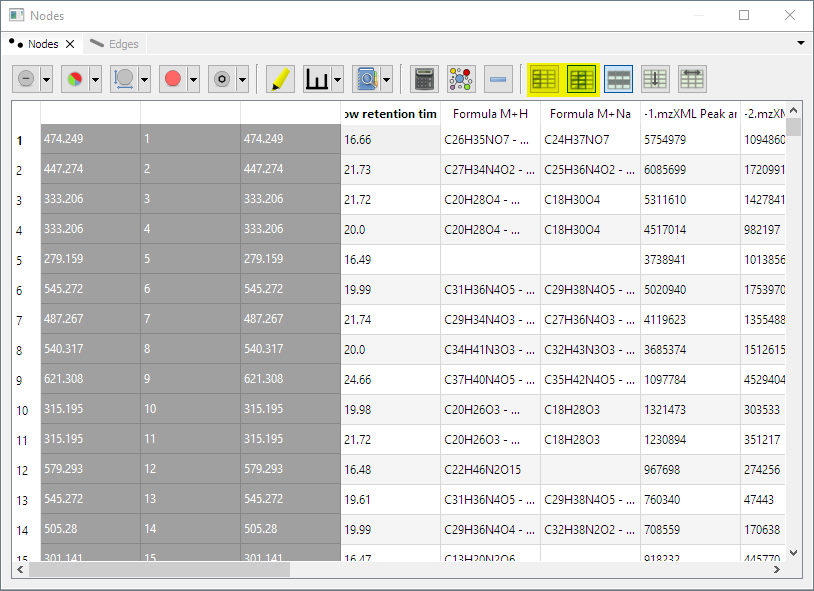
- Freeze columns feature:
- Use left mouse for selection of columns in metadata tables. Enable sorting and moving with tool button:
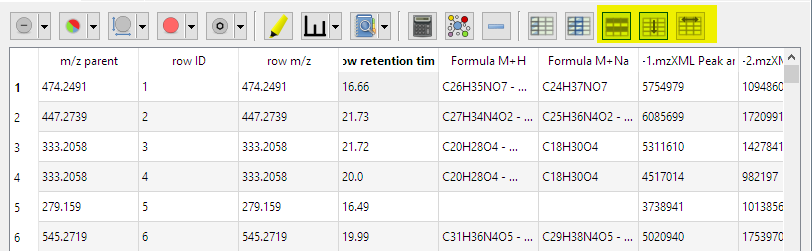
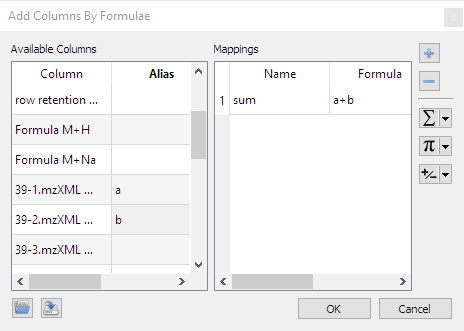
- Dialog to create new columns by combining existing ones via arithmetics formulae:
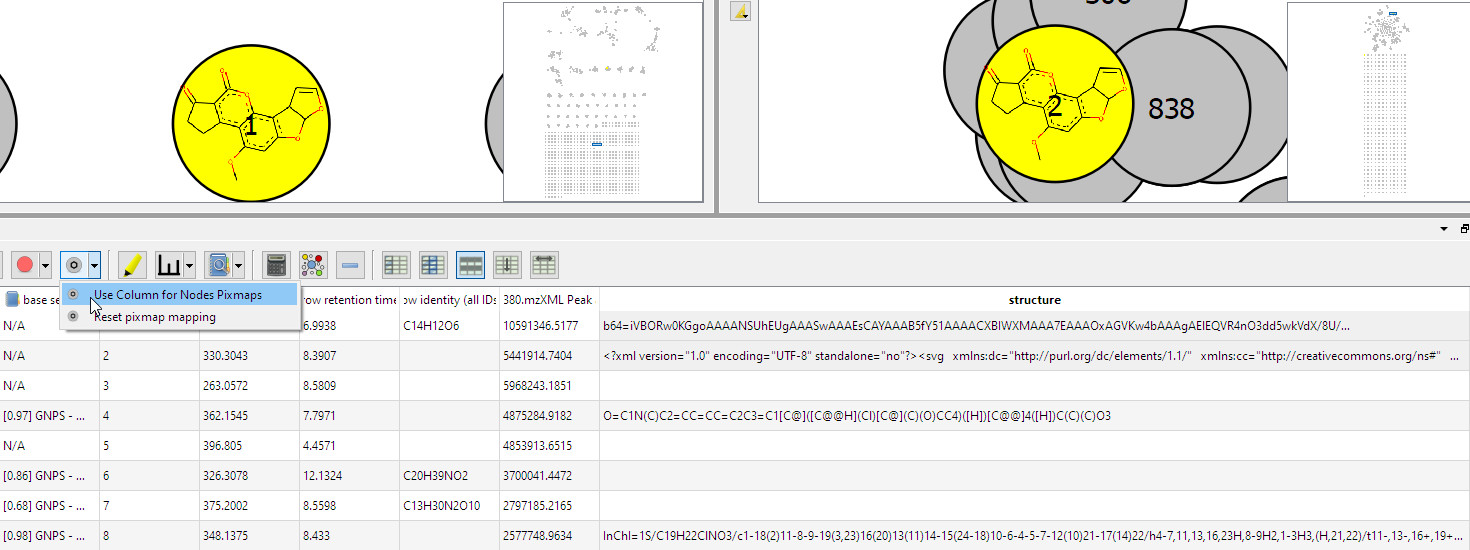
- Add pixmaps onto nodes:
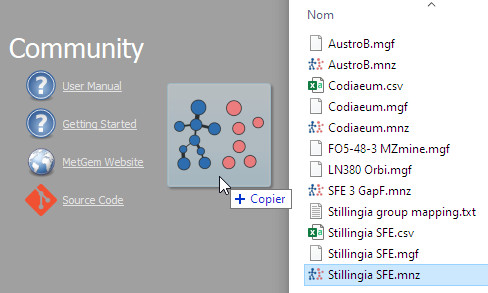
- Welcome screen visible at startup:

- You can drag and drop a project directly into the main window to open it:
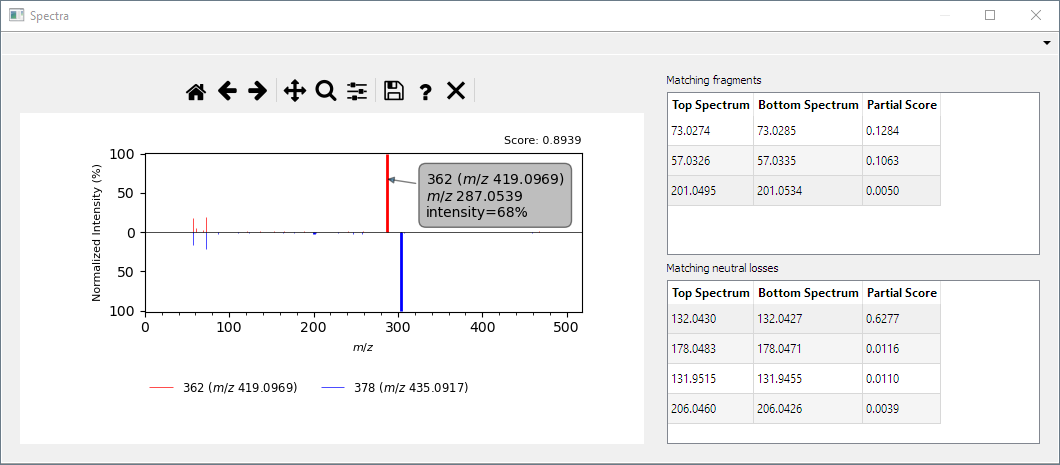
- Interface to visualize which peaks match between two spectra and cursor to show coordinates of fragments when clicked:
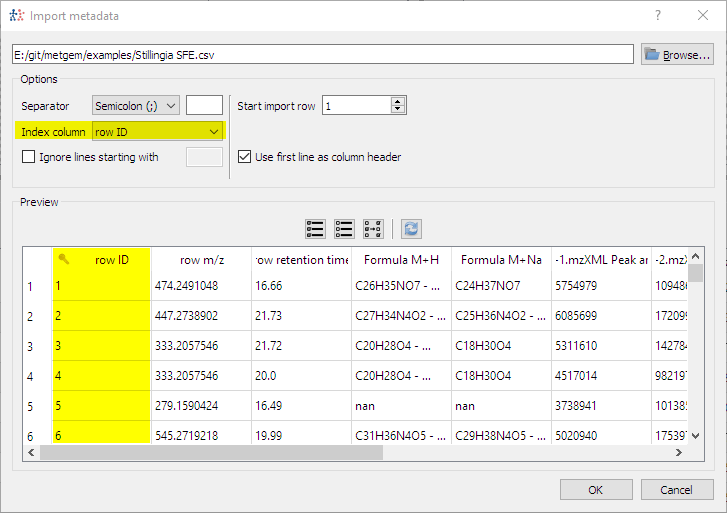
- Allow to use one column as index when importing metadata:
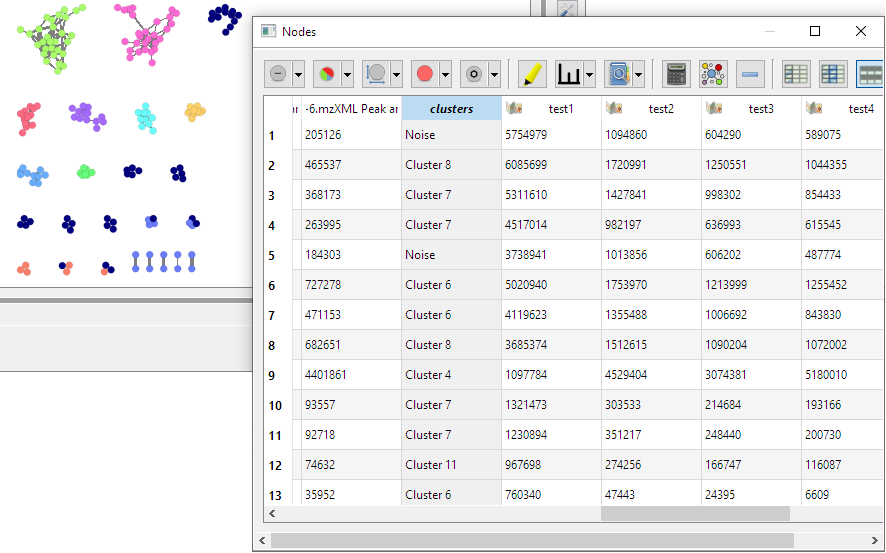
- Dialog create new column in nodes table containing name of cluster to which each node belong (using hdbscan algorithm):
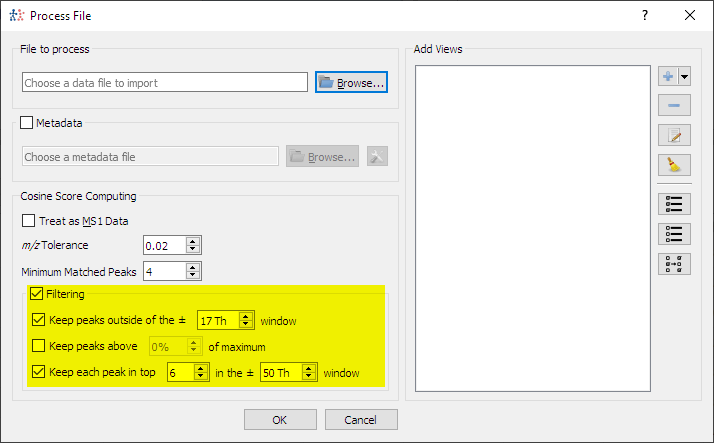
- Filtering steps can be bypassed in import dialog:
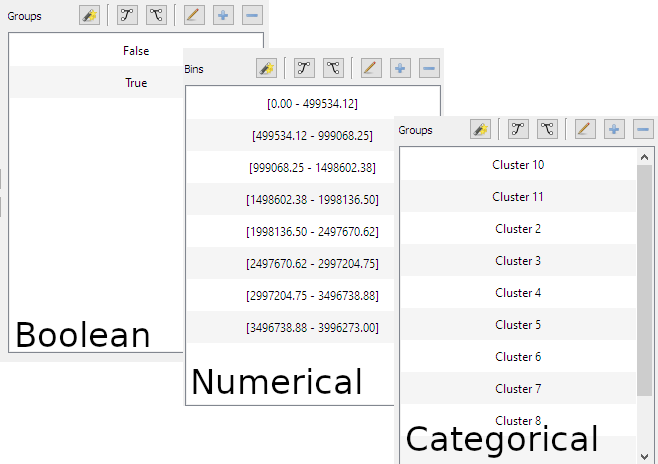
- Color Mapping Dialog can handle Boolean, Numerical or Categorical data:
A large number of bugs have also been fixed. Full changelog can be found in the release page. However, if you think you found one, feel free to fill a new issue on the github project page. You can also discuss this release in the forum.
Documentation is also available online.
Downloads are available here: