We are very excited to announce that version 1.2 of MetGem has just been released.
New features mainly include:
- Mac OS X and Linux builds
- You can now customize pie charts colors using a selection dialog:
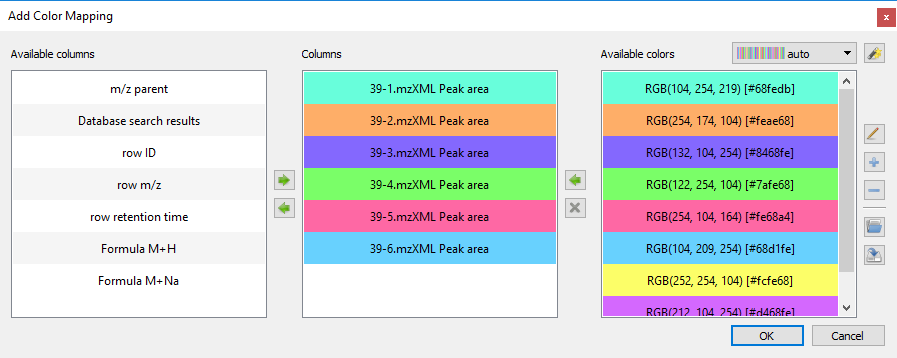
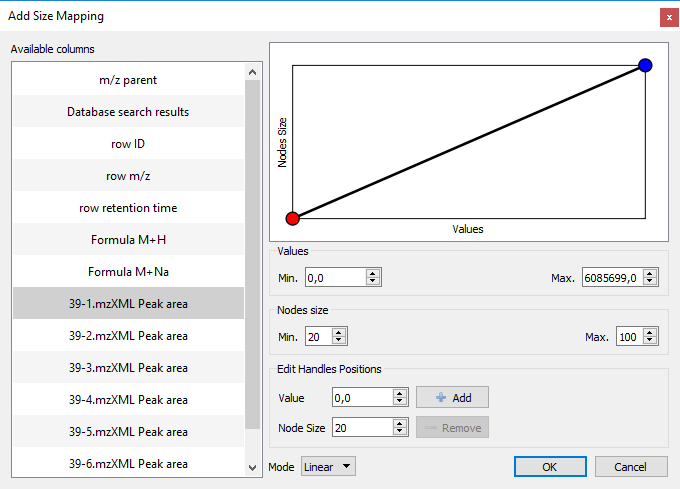
- You can now create mappings between a metadata column and sizes of nodes:
- A switch button has been added to temporarily disable pie charts
- Added a ‘lock view’ button above each view to prevent nodes to be moved by accident
- Export function for metadata and databases results
- Copy-paste function on graphs and metadata tables
- Add a new database source: MSDial
- Use of OpenGL can now be disable using a command line switch (–disable-opengl) because, on some computers, this was leading to a blank window
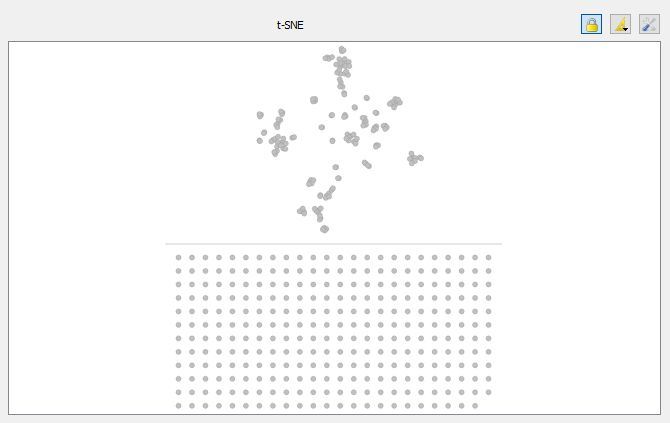
- Isolated nodes can now be hidden in both views and a line has been added to separate isolated nodes from other in the t-SNE view
- Enhanced Cytoscape export (cosine scores are exported)
- Added support for NIST format (.msp) for input data or databases
- Added a plugin system for databases sources that allows to update the download functions without updating MetGem.
A few notes about this release:
- Naming scheme for databases has changed: Delete the file spectra.sqlite in the user’s folder/databases then download all DBs again,
- To activate hide isolated nodes function for old projects on t-SNE view, please redraw the t-SNE view,
- Beware that views are now locked by default! Please click on the lock button above each view to unlock the view,
A large number of bugs have also been fixed. Full changelog can be found in the release page. However, if you think you found one, feel free to fill a new issue on the github project page.
Downloads are available here: